Cost was $399 (time limited special offer).
Exome is coding part of the Genome (essential part).
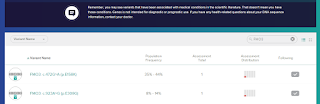
For FMO3 the test listed my 2 'wrong amino acid' variants, but not the 'silent' one.
I can't be sure it is FULL SEQUENCING, but rather a 'list of suspects'.
Overall I think it was worth it.
Best overall DNA tester I know (being both for consumer and none of the obstacles of clinical labs).
I got my exome DNA results back from GENOS.
I am very happy with the service, the process, support, and the info given.
My only gripe is I am not sure if it is FULL SEQUENCING (i.e. every codon of a gene).
It possibly is, I don't know yet.
Ways DNA labs test and give results :
1. Test for list of suspect variants/codons only (usually cheapest way).
This is no use really. Still too much unknown of the other bits. This is where they test suspect codons only. This means they choose the list of known suspects, but many more variants may yet to be discovered as fault-causing. You want to know every variant you have, not just suspects.
2. FULL SEQUENCING, but only give SELECTED variants/codons info to the person.
This means they tested every codon but they may only give what they think is relevant info on certain variants. So you still can't be sure you are getting the whole picture.
3. FULL SEQUENCING and give ALL VARIANTS whether they be thought relevant or not.
This would be as good as FULL SEQUENCING in theory I guess.
4. FULL SEQUENCING (i.e. every codon of the gene)
This is best/good, though I guess in theory you only need to know the variants, so #3 may be more practical. #3 is the same as #4 but without the 'perfect' parts (which presumably won't affect function).
I am hoping Genos is #3, but I have not found out yet. It may be #2 which would not be so good.
1. Test for list of suspect variants/codons only (usually cheapest way).
This is no use really. Still too much unknown of the other bits. This is where they test suspect codons only. This means they choose the list of known suspects, but many more variants may yet to be discovered as fault-causing. You want to know every variant you have, not just suspects.
2. FULL SEQUENCING, but only give SELECTED variants/codons info to the person.
This means they tested every codon but they may only give what they think is relevant info on certain variants. So you still can't be sure you are getting the whole picture.
3. FULL SEQUENCING and give ALL VARIANTS whether they be thought relevant or not.
This would be as good as FULL SEQUENCING in theory I guess.
4. FULL SEQUENCING (i.e. every codon of the gene)
This is best/good, though I guess in theory you only need to know the variants, so #3 may be more practical. #3 is the same as #4 but without the 'perfect' parts (which presumably won't affect function).
I am hoping Genos is #3, but I have not found out yet. It may be #2 which would not be so good.
EXOME, GENOME :
Exome = coding for all the proteins in humans
Genome = everything (Exome + Junk + promoter regions etc)
It's thought maybe 80% of faults are in the exome part.
The main reason for EXOME testing rather than GENOME is cost (at the moment).
Exome is obviously the part you can't do without. The essential part.
But people can have faults in the junk part of a gene and it can cause deficiency even though it's junk.
MY GENOS DNA RESULT
I have fully sequenced the FMO3 gene before so knew my FMO3 coding part result.
It spotted my 2 carrier copies of the common variants at codon 158 and 308.
These are reasonably common (in caucasians anyway).
There is debate if they affect function (obviously I would say they do).
Some say only if they were from same parent, or if person is homozygous (carry 2 copies).
It didn't spot a silent variant I carry. This is what makes me wonder if they only test a 'suspect list'.
Silent variants are where one of the nucleotides is wrong but the amino acid ends up the same.
FMO3 gene
532 x 3 (1596) nucleotide sequence.
makes up 532 amino acid sequence (+ stop at 533).
makes up the FMO3 protein.
Which means 532 codons where variants can be.
Clinvar list of FMO3 variants
532 x 3 (1596) nucleotide sequence.
makes up 532 amino acid sequence (+ stop at 533).
makes up the FMO3 protein.
Which means 532 codons where variants can be.
Clinvar list of FMO3 variants
IS IT A GOOD WAY TO DNA TEST FOR FMO3?
Well this is where I don't know (yet).
The best way is to have your full 532 coding sequence, or at least all the variants in that 532 coding sequence (even if they are currently deemed to not matter).
But I don't seem to have the full sequence (not that I can see so far), or all the variants (as they missed a silent one).
The give a raw file but I don't know how to open it.
Answer : I think so but I don't know.
FMO3 DATABASE
In my opinion we should get a FMO3 database going.
But how to, I don't know.
Ideally this would be both the Exon and Intron parts of FMO3 (not just the exons)
Intron is the 'non-coding' part of the gene (which could be junk + promoter region etc).
This would be a pipe dream.
My philosophy on spending on 'systemic malodors'
My philosophy is we don't know what causes it yet, so better to spend money on testing rather than spending on supplements.
FINAL COMMENT
So I can't say for sure GENOS is a great option for DNA testing, but it may be.
And at current price $399 it's cheaper than most FMO3 DNA gene testing .
Plus it's meant for the public and easy to test (saliva).
The question is how much prices will drop quickly and how well competitors do.
In the end you will probably be able to do GENOME testing for maybe $100, but when ?
Also remember, you may find results of other genes you might not want to know (e.g. prone to parkinsons, carrier of cystic fibrosis) so you need to factor in whether you want to know such things.
Note :
I am no expert, so terminology, facts may be wrong.
Links :
Genos DNA TEST LAB FOR CONSUMER
Get new posts by email

0 comments:
Post a Comment